Loading ...
Loading ...
Loading ...
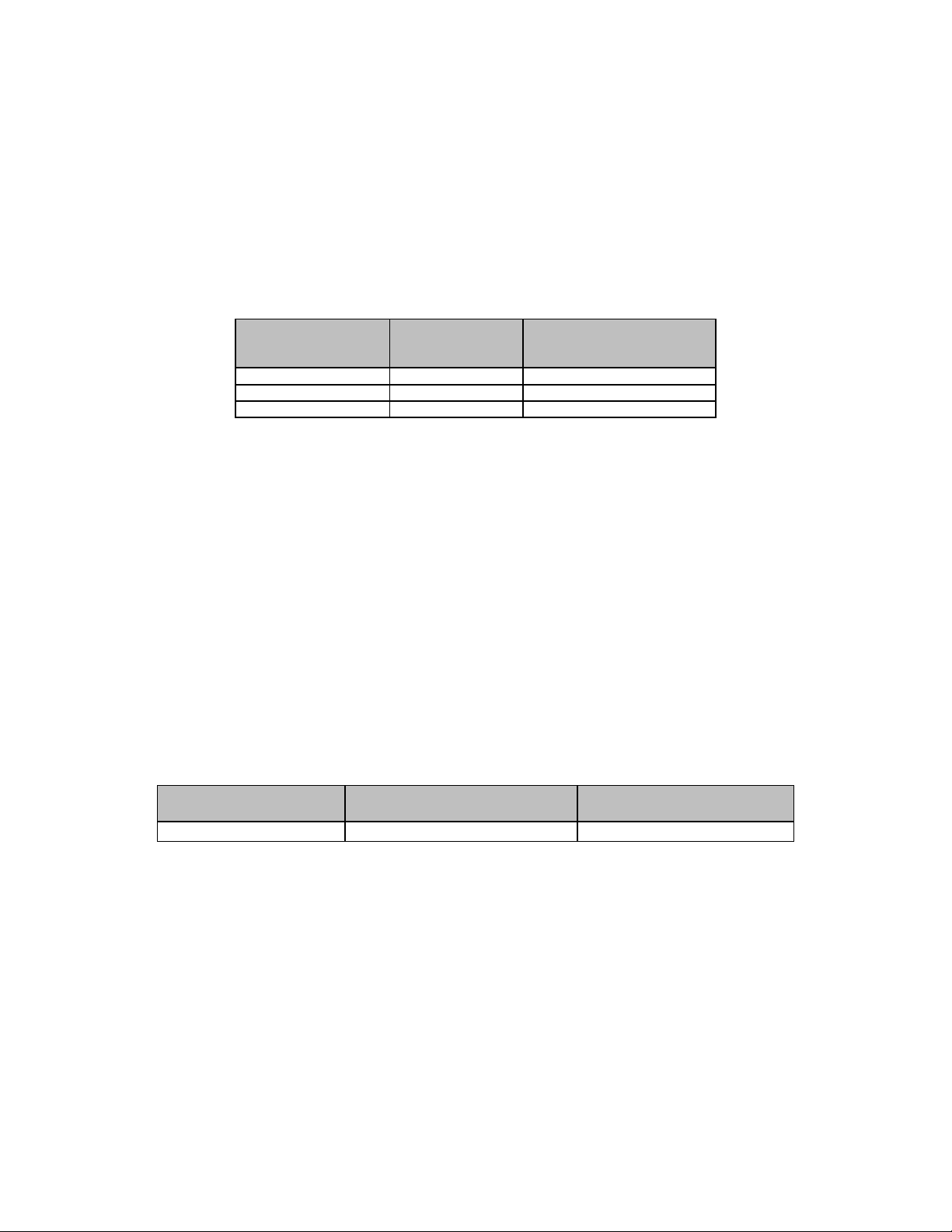
14
Clinical Study:
The performance of ID NOW COVID-19 was evaluated using contrived clinical nasopharyngeal (NP) swab
specimens obtained from individuals with signs and symptoms of respiratory illness. The samples were
prepared by spiking clinical NP swab matrix with purified viral RNA containing target sequences from the
SARS-CoV-2 genome at concentrations approximately 2x LOD and 5x LOD. Negative NP swab samples
were also tested in this study.
The table below presents ID NOW COVID-19 test agreement with the expected results by sample
concentration.
ID NOW COVID-19 Test Agreement with the Expected Results by Sample Concentration
Target
Concentration
Number
Concordant/
Number Tested
% Agreement
[95% CI]
2X LOD
20/20
100% [83.9% - 100%]
5X LOD
10/10
100% [72.3% - 100%]
Negative
30/30
100% [88.7% - 100%]
ANALYTICAL STUDIES:
Analytical Sensitivity (Limit of Detection)
ID NOW COVID-19 limit of detection (LOD) in natural nasopharyngeal swab matrix was determined by
evaluating different concentrations of purified viral RNA containing target sequences from the SARS-CoV-
2 genome.
Presumed negative natural nasopharyngeal swab specimens were eluted in ID NOW COVID-19 elution
buffer. Swab elutes were combined and mixed thoroughly to create a clinical matrix pool to be used as the
diluent. Viral RNA was diluted in this natural nasopharyngeal matrix pool to generate virus dilutions for
testing.
The LOD was determined as the lowest concentration that was detected ≥ 95% of the time (i.e.,
concentration at which at least 19 out of 20 replicates tested positive).
The confirmed LOD in natural nasopharyngeal swab matrix is presented in the table below:
Limit of Detection (LOD) Study Results
Virus
Claimed LOD
(Genome Equivalents/mL)
Positive/Replicates
SARS-CoV-2 RNA
125
19/20
Analytical Reactivity (Inclusivity)
An alignment was performed with the oligonucleotide primer and probe sequences of the ID NOW COVID-
19 assay with all publicly available SARS-CoV-2 genomic sequences submitted to NCBI Genbank, GISAID
and COG-UK databases between June 1 – June 30, 2021 to demonstrate the predicted inclusivity of the ID
NOW COVID-19 assay. A total of 26,855 complete SARS-CoV-2 sequences plus a reference genome were
submitted to NCBI GenBank and 262,260 high quality were submitted to GISAID database. The COG-UK
database contained 97,873 high quality sequences, which came from samples obtained within the same
timeframe. To avoid redundancy only the GISAID copies of any duplicated sequences were retained for
analysis bringing the total number of high quality human SARS-CoV-2 sequences available from all 3
databases to 320,634. Of the total number of sequences analyzed, 399 sequences contained at least 1 N
(unknown or unidentified nucleotide) within the target region, bringing the total number of isolates suitable
for inclusivity analysis down to 320,235. From this analysis 99.32% of the sequences provided 100%
homology to the ID NOW COVID-19 primer and probe sequences.
Loading ...
Loading ...
Loading ...